General
procedure for synthesis of curcumin conjugates and its characterization
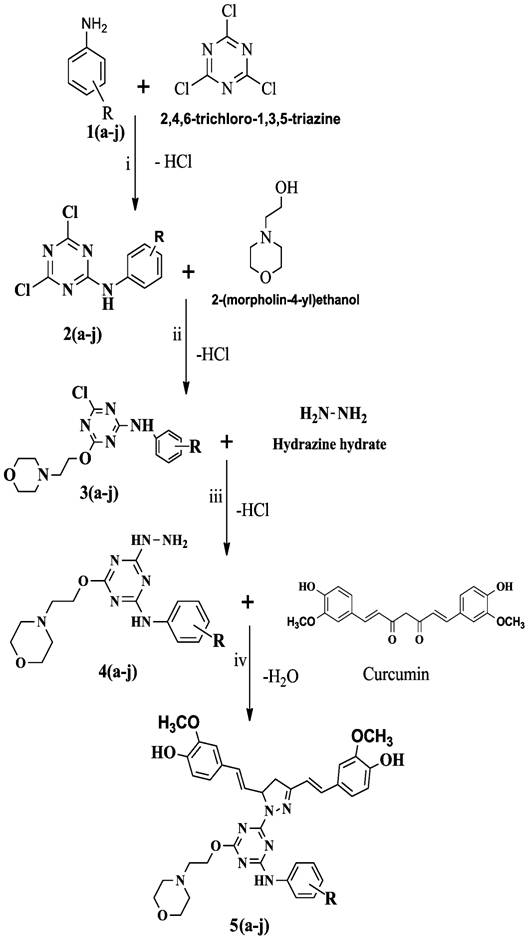
Fig.
1: Scheme
of Curcumin pyrazole triazine conjugates synthesis
Where (a-j) : a=CN.SO4,
b= CN, c = 3-F, d = 4-Br, e = 3-Cl, f = 3-NO2, g = 2-NO2,h
= 2-Cl, i = 4-NO2, j = H.
Reagents and Reaction
conditions-
(i)=
Acetone medium, Sodium bicarbonate at 0-50C.
(ii)=
Acetone medium, Sodium hydroxide at 45-500C
(iii)=
Acetone medium, sodium hydroxide at 600C
(iv)=
Glacial acetic acid, reflux temperature below 100°C
Synthesis of N substituted phenyl-1, 3,
5-triazine-2-amine derivatives as compound (2a-j)- Reaction
as shown in Fig. 1 between 2, 4, 6-trichloro-1, 3, 5-triazine (0.01M) and
1(a-j) (0.01M) was carried out by stirring in presence of (20 ml) acetone at
0-5°C. Meanwhile, (0.1M) NaHCO3
used
to neutralize liberated HCl. Completion of the reaction was verified by TLC
(acetone: benzene, 1:1). The addition of crushed ice resulted in the
precipitate and then filtered. The
purity of the compound was checked by using a melting point. The product was
air-dried [16].
Synthesis of 4-chloro-6-(2-morpholinoethoxy)-N-R-phenyl-1,3,5-triazin-2-amine
derivatives as compound(3a-j)- Equimolar amount (0.01M) of
product 2(a-j) and 2-(moroholin-4-yl) ethanol as shown in Fig. 1 was taken in a
three-necked round bottom flask fitted with the water condenser. The mixture
was refluxed at 45- 50°C in presence of acetone. Here, (0.1M) NaOH used for the
neutralization of liberated acid. Completion of the reaction was confirmed by
TLC (acetone: benzene, 2:1) and the visualization of the compound was carried
out in an iodine chamber, after that re-crystallization done by ethanol only in
the case of 4-aminobenzonitrile containing derivatives. Purity of compounds
determined by the measuring melting point, after filtration, the product was
vacuum dried [17].
Synthesis of N substituted
phenyl-4-hydrazinyl-6-(2-morpholinoethoxy)-1, 3, 5-triazin-2-amine derivatives
as compound (4a-j).
In
this step the intermediate product 3 (a-j) (0.01M) refluxed with hydrazine
hydrate (0.01M) as shown in Fig. 1 in
presence of acetone (20 ml) was taken in a three-necked round bottom flask
fitted with the water condenser. Basicity was maintained (0.1M) NaOH.
Completion of the reaction was monitored by TLC with appropriate solvent
systems. The visualization of the compound was carried out in an iodine
chamber. The product was purified by re-crystallization by using ethanol in the
case of 4-aminobenzonitrile containing triazine. Thereafter compound was
filtered and vacuum dried.
Synthesis of of N-phenylamino -1, 3,
5-triazin-2-yl)-4, 5-dihydro-1H-pyrazol-3-yl) vinyl)-2-methoxyphenol
derivatives as compound 5(a-j)- In the final step as shown
in Fig. 1 above product 4(a-j) (0.01M) was conjugated to
1,7-bis(4-hydroxy-3-methoxyphenyl) hepta-1,6-diene-3,5-dione (curcumin,0.01M)
at reflux temperature in presence of glacial acetic acid (30 ml). Completion of
the reaction was monitored by the TLC (acetone: benzene, 1:1). The
visualization of the compound was carried out in an iodine chamber. The product
was re-crystallized with ethanol only in the case of 4-aminobenzonitrile
containing triazine. After filtration of the compound, it was dried in a
vacuum. The purity of the compound was checked by determining the melting point
[18].
Antibacterial activity- Different
conjugates were screened for antibacterial activity according to the guidelines
of the Clinical Laboratory Standard Institute (CLSI). Different pathogenic
strains of gram-negative bacteria and gram-positive bacteria were used for
screening. The experiment was conducted using nutrient broth media. The
nutrient broth culture with loopful bacterial strains was incubated at 37+1°C for 16–18 hrs, and the microbial culture was
adjusted to the McFarland standard. Diluting the bacterial suspension with
sterile solution yielded a final concentration of 1.5x108 CFU/mL.
The plates of nutrient agar media were prepared. With the help of stainless
steel cork, 5-mm diameter wells were made into swabbed agar plates. To make a
neat solution, testing compounds were dissolved in dimethyl sulfoxide (DMSO).
Following that, the wells were loaded with 30 µl of testing samples and
incubated at 37+1°C for 16–18 hrs and it was performed in triplicate.
Their activity was assessed by measuring the zone of inhibition against
bacterial pathogens using a zone reader (Himedia zone scale) [19].
Minimum inhibitory concentration (MIC) and
Minimum bactericidal concentration (MBC)- Different bacterial strains
were screened for MIC. Each well of a 96-well microplate was filled with 75 µl
nutrient broth and for testing drugs of many concentrations in 2-fold dilution
as 1024, 512, 256, 128, 64, 32, 16 µg/ml. Then 75 µl of each test organism was
inserted in each well. The remaining two wells in each row of microplates were
then allowed to go negative control (i.e., the extract was substituted with 50
µl of 10% DMSO) and positive control as no extract but 50 µl antibiotic.
Finally, each well-received 50 µl of resazurin solution. The experiments were
conducted in triplets and bacterial isolates were incubated for 24 h at 37+1°C.
The last volume of a drug capable of penetrating visible microbial growth of
microorganisms or no change in resazurin dye colour was then reported as the
MIC value [20]. To assess MBC, a loopful of the mixture of each well
that did not exhibit microbial growth was sub-cultured by streaking on nutrient
agar plates and incubated for 24 h at 37+1°C. MBC was recorded as the
lowest concentrations of extract that did not display any established colony [21].
Antifungal activity- This
method was performed according to the guidelines of CLSI. Sabouraud dextrose
agar/broth media for C. albicans and potato dextrose agar for A.
fumigatus were used to experiment. Broth culture with a loopful fungal
strain was incubated at 37+1°C for 16–18 hrs for yeast and 28+1°C
for 72 hrs mold and microbial culture was adjusted to McFarland standard for
yeast 1.5 x 108 and spore suspension adjusted for mold at 5x104
spores. Total 20 ml media were poured then solidifying plates were
swabbed using a sterilized cotton swab with 100 µl. With the help of a cork
borer of 5 mm diameter wells were made. Testing compounds were dissolved in
DMSO to make a neat solution. After that the wells were loaded with 30 µl of
testing samples and allowed to incubate at 37+1°C for 16–18 hrs for
yeast and 28+1°C for 72 hrs mold. Their activity was evaluated by
measuring the zone of inhibition. The procedure was performed in triplicate for
the pathogen. The compound was tested against the test organism in triplicate [19].
Statistical
Analysis- The antibacterial activity of different curcumin
conjugates synthesized in the present study was analyzed using two-way Analysis
of Variance (ANOVA) followed by F-test and the significance was tested at 5%,
1% and 0.1% and results interpreted accordingly.
RESULT
Chemistry and
characterization of synthesized compounds- The
designed library of target compounds and respective intermediates were
synthesized as outlined in Fig. 1. Aromatic amines (4-aminobenzonitrile, 4-nitroaniline,
3-nitroaniline, 3-floroaniline, 4-bromoaniline, 3-chloroaniline, 2-niroaniline,
2-chloroaniline, and aniline) and 2, 4, 6, - trichloro-s-triazine are the
reactants involved in the first step in the scheme for the synthesis of
curcumin conjugates. The formation of mono substituted triazine occurred as a
result of a nucleophilic aromatic substitution reaction with the hydrolysis of
one of the chloro groups in the form of HCl in the presence of acetone as a
solvent and sodium bicarbonate (NaHCO3) as a neutralizing agent. In
the second step an intermediate 2(a-j) reacted with 2-(morpholin-4-yl) ethanol,
where OH group of morpholine hydrolyzed with one of the chloro group via
nucleophilic substitution reaction of s-triazine to form di-substituted
s-triazine derivative. This reaction took place in the presence of acetone and
liberated acid (HCl), which was neutralized by sodium hydroxide (NaOH). The
second intermediate 3(a-j) of the next step, then reacted with hydrazine
hydrate to form tri substituted 1, 3, 5-triazine with HCl liberation. In the last
step, curcumin conjugated to 4(a-j) intermediate that resulted in the
cyclization of diketone moiety of curcumin by hydrazine hydrate with the
liberation of a water molecule to form final product as
N-phenyl-4-hydrazinyl-6-)2- morpholinoethoxy)-1,3,5-triazine-2-amine
derivatives. The final compounds were characterized by various spectroscopic
data’s such as FT-IR, 1H NMR, 13C NMR and Mass spectra.
4-((4-(3,5-bis(4-hydroxy-3-methoxyphenethyl)-4,5-dihydro-1Hpyrazol-1-yl)-6
(2morpholinoethoxy)-1,3,5-triazin-2-yl)amino)benzonitrile sulfate hydrate. A2- Yellow
amorphous solid; Yield: 40%; M.P.: 2100C FTIR (νmax; cm-1
KBr): 3196.38 (O-H stretching), 3080.37 (N-H stretching), 2923.38 (Aromatic C-H
stretching), 2230.42 (OCH3 stretching), 1738.28 (C=O stretching),
1683.31 (C=N stretching), 1586.20 (CH2
bending), 1551.21 (C=C stretching), 1367.27 (N-N stretching), 779.42; 1H NMR (400MHz, DMSO-d6, TMS) δ ppm: 10.84 (s, 2H,
Ar-OHx2), 7.46 (d, 2H, J=0.01 Hz,Ar-H), 7.32 (d, 2H, J=0.71 Hz, Ar-H), 6.81 (d,
1H, J=0.78 Hz, pyrazole-H), 6.77 (d, 2H, J=0.01 Hz, Ar-H), 6.73 (d, 2H, J=0.78
Hz, Ar-H), 6.57 (d, 2H, J=0.72 Hz, Ar-H), 3.83 (s, 1H, NH), 3.68 (s, 6H, OCH3x2),
3.60 (t, 2H, J=3.47 Hz, CH2, methylene), 3.58 (d, 4H,
J=0.14 Hz, morpholine-H), 2.51-1.91 (m, 4H, aliphatic CHx4), 2.49 (t, 2H,
J=3.60 Hz, CH2, methylene), 2.50 (d, 4H, J=3.60 Hz, morpholine-H); 13C
NMR (100MHz, DMSO-d6) δ ppm:172.07, 165.52, 164.39, 161.36, 148.19,
133.07, 132.73, 124.07, 120.57, 119.04, 113.28, 108.87, 108.73, 107.86, 104.95,
40.03, 39.82, 39.62, 39.41, 39.20, 38.99, 38.78, 21.40, 21.04; GC-MS: 281.0
(Triazine-Morpholine-M), 326.5 (Triazine- morpholine and amine, M+H).
4-((4-(3,5-bis(4-hydroxy-3-methoxyphenethyl)-4,5-dihydro-1Hpyrazol-1-yl)-6-(2-morpholinoethoxy)-1,3,5-triazin-2-yl)amino)benzonitrile.
A2- Dark
brown amorphous solid; Yield: 44%; M.P.:1800C; FTIR (νmax; cm-1
KBr): 3196.38 (O-H stretching), 3080.37 (N-H stretching), 2923.38 (Aromatic C-H
stretching), 2230.42 (OCH3 stretching), 1738.28 (C=O stretching),
1683.31 (C=N stretching), 1586.20 (CH2
bending), 1551.21 (C=C stretching), 1367.27 (N-N stretching), 779.42; 1H NMR (400MHz, DMSO-d6, TMS) δ ppm: 10.84 (s, 2H,
Ar-OHx2), 7.46 (d, 2H, J=0.01 Hz,Ar-H), 7.32 (d, 2H, J=0.71 Hz, Ar-H), 6.81 (d,
1H, J=0.78 Hz, pyrazole-H), 6.77 (d, 2H, J=0.01 Hz, Ar-H), 6.73 (d, 2H, J=0.78
Hz, Ar-H), 6.57 (d, 2H, J=0.72 Hz, Ar-H), 3.83 (s, 1H, NH), 3.68 (s, 6H, OCH3x2),
3.60 (t, 2H, J=3.47 Hz, CH2, methylene), 3.58 (d, 4H,
J=0.14 Hz, morpholine-H), 2.51-1.91 (m, 4H, aliphatic CHx4), 2.49 (t, 2H,
J=3.60 Hz, CH2, methylene), 2.50 (d, 4H, J=3.60 Hz, morpholine-H); 13C
NMR (100MHz, DMSO-d6) δ ppm:172.07, 165.52, 164.39, 161.36, 148.19,
133.07, 132.73, 124.07, 120.57, 119.04, 113.28, 108.87, 108.73, 107.86, 104.95,
40.03, 39.82, 39.62, 39.41, 39.20, 38.99, 38.78, 21.40, 21.04; GC-MS: 281.0
(Triazine-Morpholine-M), 326.5 (Triazine- morpholine and amine, M+H).
4-(2-(1-(4-((4-f luorophenyl)amino)-6-(2- morpholinoethoxy)-1,3,5-triazin-2-yl)-3(4-hydroxy-3-methoxy
styryl)-4,5-dihydro-1Hpyrazol-5-yl)ethyl)-2-methoxyphenol. A3- Dark
brown amorphous solid; Yield: 67.05%; M.P.:580C; FTIR (νmax; cm-1
KBr): 3367.80 (O-H stretching), 1579.51 (C=N stretching), 1506.50 (CH2 bending), 1395.27 (N-N stretching), 1262.78,
1124.04 (C-F stretching), 906.25, 608.10; 1H
NMR (400MHz, DMSO-d6, TMS) δ ppm: 8.08 (s, 2H, Ar-OHx2), 7.63 (d, 2H,
J=1.43 Hz,Ar-H), 7.46 (d, 2H, J=1.55 Hz, Ar-H), 7.14 (d, 1H, J=1.42 Hz, Ar-H),
7.04 (d, 2H, J=0.72 Hz, Ar-H), 7.00 (d, 2H, J=0.72 Hz, Ar-H), 6.97 (t, 2H,
J=3.69 Hz, CHx-2), 6.95 (t, 2H, J=0.72 Hz, CHx-2), 6.70 (s, 1H,pyrazole-H),
4.08 (t, 2H, J=3.86 Hz, CH2, methylene), 3.83 (s, 1H,
NH), 3.81 (s, 6H, OCH3x2), 3.57-2.50 (m, 8H, morpholine-H), 2.53 (t,
2H, J=15.53 Hz, CH2, methylene); 13C NMR (100MHz, DMSO-d6) δ ppm:78.90, 78.11, 78.57,
78.24, 66.11, 60.91, 60.47, 57.99, 56.46, 53.53, 53.32, 40.12, 39.91, 39.70,
39.49, 39.28, 39.07, 38.86, 21.40, 20.61; GC-MS: 281.0 (Triazine-Morpholine-M),
355.1 (Triazine- morpholine and amine, M+2H).
4-(2-(1-(4-((4-bromophenyl)amino)-6-(2-morpholinoethoxy)-1,3,5-triazin-2-yl)-3-(4-hydroxy-3-methoxystyryl)-4,5-dihydro-1Hpyrazol-5-yl)ethyl)-2-methoxyphenol.
A4- Dark
brown amorphous solid; Yield: 66.07%; M.P.:840C; FTIR (νmax; cm-1
KBr): 3321.49 (O-H stretching), 1567.89 (C=N stretching), 1510.78 (CH2 bending), 1396.32 (C-Br stretching),
1256.03, 1019.69, 905.06; 1H NMR (400MHz, DMSO-d6, TMS)
δ ppm: 8.19 (s, 2H, Ar-OHx2), 7.44 (d, 2H, J=0.32 Hz,Ar-H), 7.40 (d, 2H,
J=0.34 Hz, Ar-H), 7.38 (d, 1H, J=0.70 Hz, Ar-H), 6.82 (d, 2H, J=0.34 Hz, Ar-H),
6.80 (d, 2H, J=0.05 Hz, Ar-H), 6.77 (t, 2H, J=0.32 Hz, CHx-2), 6.74 (t, 2H,
J=0.34 Hz, CHx-2), 6.68 (s, 1H,pyrazole-H), 4.10 (t, 2H, J=3.86 Hz, CH2, methylene), 3.92 (s, 1H, NH), 3.82 (s, 6H,
OCH3x2), 3.58-2.51 (m, 8H, morpholine-H), 2.54 (t, 2H, J=3.01 Hz, CH2,
methylene); 13C NMR (100MHz, DMSO-d6) δ ppm:79.06, 78.73, 78.41,
66.05, 60.49, 57.99, 53.55, 40.17, 39.96, 39.75, 39.51, 39.33, 39.13, 38.92,
21.25; GC-MS: 281.0 (Triazine-Morpholine-M), 417.3 (Triazine- morpholine and
amine, M+3H), 647.5 (Curcumin-Para bromoanilineM+Cl).
4-(2-(1-(4-((3-chlorophenyl)amino)-6-(2-morpholinoethoxy)-1,3,5-triazin-2-yl)-3-(4-hydroxy-3-methoxy
styryl)-4,5-dihydro-1Hpyrazol-5-yl)ethyl)-2-methoxyphenol. A5- Dark
brown amorphous solid; Yield: 32.67%; M.P.:800C; FTIR (νmax; cm-1
KBr): 3308 (O-H streching), 2924 (C-H
streching), 1643 (C=C streching), 1395 (CH3), 1395 (CH3
vibration), 1340 (C-H alkane stretching), 1127, 1021 (C-O streching), 1095
(C-O-C), 905 (CH= CH2), 671 (C-Cl), 538; 1H
NMR (400MHz, DMSO, TMS) δ ppm: 8.19 (s, 2H, Ar-OHx2), 8.04 (s, 1H, Ar-NH),
7.25 (d, 2H, J=1.64 Hz, Ar-H), 7.23 (d,
1H, J=2.7 Hz, Ar-H), 7.22 (d, 1H, J=2.3 Hz, Ar-H), 7.04 (d, 1H, J=1.7 Hz,
Ar-H), 7.03 (d, 1H, J=4.6 Hz, Ar-H), 7.01 (d, 1H, J=1.4 Hz, Ar-H), 6.97 (d, 1H,
J=4.6 Hz, Ar-H), 6.95 (d, 1H, pyrazole-H), 6.78 (d, 1H, J=1.5 Hz, Ar-H), 6.66
(d, 2H, J=4.6 Hz, Ar-H), 6.63 (d, 1H, J=1.2 Hz, Ar-H), 4.18 (s, 2H, CH2, Methylene), 3.85-3.76 (s, 6H,2xOCH3),
2.72 (s, 2H, CH2, Methylene), 3.57-2.51 (m,
8H, 4x CH2, Morpholine-H), 2.53 (s,
2H, CH2 , Methylene); 13C
NMR (400MHz, DMSO) δ ppm:79.36, 78.93, 78.73, 78.43, 66.14, 59.13, 53.62,
43.12, 39.96, 39.75, 39.55, 39.34, 39.13, 38.92; GC-MS: 131.0 (Morpholine-M),
253 (Triazine- amine, M+H), 377 (Curcumin, M+Cl).
4-(2-(5-(4-hydroxy-3-methoxyphenethyl)-1-(4-(2-morpholinoethoxy)-6-((3
nitrophenyl)amino)-1,3,5-triazin-2-yl)-4,5-dihydro-1H-pyrazol-3-yl)vinyl)-2-methoxyphenol.
A6- Dark
brown amorphous solid; Yield: 93%; M.P.:800C; FTIR (νmax; cm-1
KBr): 3372.90 (O-H stretching), 1580.12 (C=N stretching), 1519.89 (NO2
stretching), 1396.14 (CH3
vibration), 1342.84 (N-N stretching),
1256.92, 1093.49, 905.63; 1H NMR
(400MHz, DMSO-d6, TMS) δ ppm: 8.19 (s, 2H, Ar-OHx2), 7.51 (d, 1H, J=1.00
Hz,Ar-H), 7.13 (d, 1H, J=1.17 Hz, Ar-H), 7.12 (d, 1H, J=1.16 Hz, Ar-H), 7.10
(d, 1H, J=2.58 Hz, Ar-H), pyrazole-H),
6.79 (d, 2H, J=11.18 Hz, Ar-H), 6.63 (d, 2H, J=0.86 Hz, Ar-H), 6.54 (d, 2H,
J=1.16 Hz, Ar-H), 6.53 (t, 4H, J=2.58
Hz, CHx-4), 6.51 (s, 1H,pyrazole-H), 4.11 (t, 2H, J=12.07 Hz, CH2), 3.99 (s, 1H, NH), 3.83 (s, 6H, OCH3x2),
3.56-2.52 (m, 8H, morpholine-H), 2.55 (t, 2H, J=3.01 Hz, CH2); 13C NMR (100MHz, DMSO-d6) δ ppm:79.06, 78.93, 78.72,
78.04, 66.12, 66.07, 60.50, 58.01, 53.55, 53.34, 40.16, 39.95, 39.74, 39.53,
39.33, 39.12, 38.91, 21.07; GC-MS: 281.0 (Triazine-Morpholine-M), 346.2
(Triazine- morpholine and amine, M+H), 617.3 (Curcumin-meta nitroanilineM+Cl).
4-((4-(3,5-bis(4-hydroxy-3-methoxyphenethyl)-4,5-dihydro-1Hpyrazol-1-yl)-6-(2
morpholinoethoxy)-1,3,5-triazin-2-yl)amino)benzonitrile sulfate hydrate. A7- Yellow
amorphous solid; Yield: 40%; M.P.: 2100C FTIR (νmax; cm-1
KBr): 3196.38 (O-H stretching), 3080.37 (N-H stretching), 2923.38 (Aromatic C-H
stretching), 2230.42 (OCH3 stretching), 1738.28 (C=O stretching),
1683.31 (C=N stretching), 1586.20 (CH2
bending), 1551.21 (C=C stretching), 1367.27 (N-N stretching), 779.42; 1H NMR (400MHz, DMSO-d6, TMS) δ ppm: 10.84 (s, 2H,
Ar-OHx2), 7.46 (d, 2H, J=0.01 Hz,Ar-H), 7.32 (d, 2H, J=0.71 Hz, Ar-H), 6.81 (d,
1H, J=0.78 Hz, pyrazole-H), 6.77 (d, 2H, J=0.01 Hz, Ar-H), 6.73 (d, 2H, J=0.78
Hz, Ar-H), 6.57 (d, 2H, J=0.72 Hz, Ar-H), 3.83 (s, 1H, NH), 3.68 (s, 6H, OCH3x2),
3.60 (t, 2H, J=3.47 Hz, CH2, methylene), 3.58 (d, 4H,
J=0.14 Hz, morpholine-H), 2.51-1.91 (m, 4H, aliphatic CHx4), 2.49 (t, 2H,
J=3.60 Hz, CH2, methylene), 2.50 (d, 4H, J=3.60 Hz, morpholine-H); 13C
NMR (100MHz, DMSO-d6) δ ppm:172.07, 165.52, 164.39, 161.36, 148.19,
133.07, 132.73, 124.07, 120.57, 119.04, 113.28, 108.87, 108.73, 107.86, 104.95,
40.03, 39.82, 39.62, 39.41, 39.20, 38.99, 38.78, 21.40, 21.04; GC-MS: 281.0
(Triazine-Morpholine-M), 326.5 (Triazine- morpholine and amine, M+H).
4-(2-(1-(4-((2-chlorophenyl)amino)-6-(2-morpholinoethoxy)-1,3,5-triazin-2-yl)-3-(4-hydroxy-3-methoxystyryl)-4,5-dihydro-1Hpyrazol-5-yl)ethyl)-2-methoxyphenol.
A8- Dark
brown amorphous solid; Yield: 81.15 %; M.P.:1500C; FTIR (νmax;
cm-1 KBr): 3371.84 (O-H stretching), 1562.62 (C=N stretching),
1393.29 (CH3 vibration), 1260.51 (N-N stretching), 1125.11, 1092.48, 749.94 (C-Cl stretching); 1H NMR (400MHz, DMSO-d6, TMS) δ ppm: 8.17 (s, 2H,
Ar-OHx2), 7.44 (d, 1H, J=0.76 Hz,Ar-H), 7.42 (d, 1H, J=0.89 Hz, Ar-H), 7.32 (d,
1H, J=1.36 Hz, Ar-H), 7.31 (d, 1H, J=0.89 Hz, Ar-H), 7.16 (d, 2H, J=5.19 Hz,
Ar-H), 7.11 (d, 2H, J=1.36 Hz, Ar-H), 6.79 (d, 2H, J=0.89 Hz, Ar-H), 6.76-6.67 (t, 4H, J=5.19 Hz, CHx-4), 6.63 (s,
1H,pyrazole-H), 4.11 (t, 2H, J=5.24 Hz, CH2),
3.86 (s, 1H, NH), 3.82 (s, 6H, OCH3x2), 3.57-2.52 (m, 8H,
morpholine-H), 2.56 (t, 2H, J=11.38 Hz, CH2); 13C
NMR (100MHz, DMSO-d6) δ ppm: 129.07, 127.05, 79.02, 78.89, 78.69, 78.36,
66.07, 60.50, 58.00, 53.55, 53.34, 40.17, 39.96, 39.75, 39.54, 39.33, 39.12,
38.92, 21.42; GC-MS: 282.7 (Triazine-Morpholine-M+H), 601.2 (Curcumin-Para
chloroanilineM+Cl), 697.2 (M).
4-(2-(5-(4-hydroxy-3-methoxyphenethyl)-1-(4-(2-morpholinoethoxy)-6-((4
nitrophenyl) amino)-1,3,5-triazin-2-yl)-4,5-dihydro-1H-pyrazol-3-yl)vinyl)-2-methoxyphenol.
A9- Dark
brown amorphous solid; Yield: 71%; M.P.: 600C; FTIR (νmax; cm-1
KBr): 3246.17 (O-H stretching), 2933.11 (C-H stretching), 1729.71 (C=O
stretching), 1568.63 (C=N stretching), 1498.61 (NO2 stretching), 1378.73 (CH3 vibration), 1323.84
(N-N stretching), 1253.52, 1023.08,
905.28; 1H NMR (400MHz, DMSO-d6, TMS)
δ ppm: 8.17 (s, 2H, Ar-OHx2), 8.06 (d, 2H, J=4.91 Hz,Ar-H), 7.53 (d, 2H,
J=1.27 Hz, Ar-H), 7.18 (d, 2H, J=2.62 Hz, Ar-H), 7.09 (d, 2H, J=2.61 Hz, Ar-H),
7.00 (d, 2H, J=1.27 Hz, Ar-H), 6.97 (t, 2H, J=4.25 Hz, CHx2), 6.95 (t, 2H,
J=2.62 Hz, CHx2), 6.67 (s,
1H,pyrazole-H), 4.34 (t, 2H, J=2.21 Hz, CH2),
3.92 (s, 1H, NH), 3.84 (s, 6H, OCH3x2), 3.70-2.55 (m, 8H,
morpholine-H), 2.44 (t, 2H, J=3.85 Hz, CH2); 13C NMR
(100MHz, DMSO-d6) δ ppm:78.81, 78.48, 78.16, 39.87, 39.67, 39.46; GC-MS:
281.1 (Triazine-Morpholine-M), 345.2 (Triazine- morpholine and amine, M).
4-(2-(5-(4-hydroxy-3-methoxyphenethyl)-1-(4-(2-morpholinoethoxy)-6-(phenylamino)-1,3,5-triazin-2-yl)-4,5-dihydro-1H-pyrazol-3-yl)vinyl)-2-methoxyphenol.
A10- Dark brown amorphous solid; Yield: 68.49%; M.P.:1600C;
FTIR (νmax; cm-1 KBr): 3032.76 (C-H stretching), 1720.60 (C=O
stretching), 1551.29 (C=N stretching), 1389.86 (CH3 vibration),
1324.82 (N-N stretching), 1264.18,
1022.77, 958.75; 1H NMR (400MHz, DMSO-d6, TMS)
δ ppm: 9.65 (s, 2H, Ar-OHx2), 7.62 (d, 2H, J=1.10 Hz,Ar-H), 7.31 (d, 2H,
J=4.65 Hz, Ar-H), 7.14 (d, 2H, J=1.87 Hz, Ar-H), 7.10 (d, 2H, J=5.70 Hz, Ar-H),
7.07 (d, 1H, J=5.06 Hz, Ar-H), 6.87 (d, 2H, J=2.26 Hz, Ar-H), 6.86 (t, 2H,
J=2.17 Hz, CHx2), 6.84 (t, 2H, J=1.09 Hz, CHx2), 6.68 (s, 1H, pyrazole-H), 5.87 (t, 2H, J=1.87
Hz, CH2), 3.84 (s, 1H, NH), 3.35
(s, 6H, OCH3x2), 3.56-2.49 (m, 8H, morpholine-H), 2.51 (t, 2H,
J=1.45 Hz, CH2); 13C
NMR (100MHz, DMSO-d6) δ ppm:183.17, 165.57, 164.42, 161.40, 154.21,
149.33, 148.23, 147.96, 140.66, 137.28, 128.74, 126.30, 124.11, 121.17, 121.06,
115.67, 111.32, 107.93, 55.67, 40.12, 39.91, 39.08, 38.87, 21.48; GC-MS: 281.1
(Triazine-Morpholine-M), 537.0 (Curcumin-triazine-aniline M+3H).
Antibacterial activity of synthesized
curcumin derivatives- The
antibacterial activity of ten Curcumin pyrazole triazine conjugates was
evaluated and the screening results are reported in Table 1. It was evident from
the assay that compound 5 and 8 both have chlorine atom in their aniline moiety
enhanced their antimicrobial activity. In the case of the most potent drug A5
it has been reported that among gram-positive bacteria B. subtilis, S. aureus and C. perfringens
were found to have the highest zone of inhibition ranged 27. Total 26 mm and 25
mm, among gram-negative bacteria drug was found most potent against P. aeruginosa
with a zone of inhibition of 28 mm. The overall efficacy of different compounds
can be found as 5>8>3>4>6>7>10>1>2>9.
Table 1: Antibacterial activity of curcumin pyrazole triazine conjugates
|
Test organisms |
Zone of inhibition (mm diameter) |
||||||||||
|
A1 |
A2 |
A3 |
A4 |
A5 |
A6 |
A7 |
A8 |
A9 |
A10 |
||
|
1 |
L. monocytogenes |
5.66 |
7.33 |
14.66 |
14.66 |
13.66 |
15.00 |
6.33 |
14.00 |
7 |
8.33 |
|
2 |
B. cereus |
8.66 |
8.33 |
13.33 |
11.33 |
12.33 |
10.33 |
11.33 |
20.33 |
7.33 |
10.33 |
|
3 |
B. subtilis |
8.33 |
10.33 |
15.66 |
27.66 |
26.33 |
22.66 |
17.66 |
26.67 |
7.33 |
9.00 |
|
4 |
C. perfringens |
11.00 |
10 |
19.66 |
16.66 |
24.66 |
20.66 |
11.33 |
15.67 |
9.66 |
11.67 |
|
5 |
S. aureus |
7.00 |
8.33 |
31.66 |
27.66 |
26.33 |
23.66 |
13.66 |
31.33 |
7.66 |
10.33 |
|
6 |
S. pyogenes |
5.66 |
6.33 |
6.66 |
0 |
11.33 |
0 |
8.00 |
13.33 |
0 |
6.33 |
|
7 |
E.
coli |
6.00 |
6 |
20.00 |
18.66 |
17.66 |
16.66 |
11.66 |
14.67 |
6.33 |
6.00 |
|
8 |
P. aeruginosa |
0 |
0 |
22.66 |
26.66 |
27.66 |
25.66 |
11.66 |
25.67 |
0 |
7.00 |
|
9 |
S. typhi |
6.33 |
6.33 |
14.33 |
16.66 |
14.66 |
17.00 |
11.33 |
17.67 |
6 |
6.00 |
|
10 |
S. dysenteriae |
6.33 |
6.66 |
15.66 |
16.00 |
15.33 |
16.33 |
11.66 |
17.33 |
7.66 |
6.66 |
|
11 |
V. cholera |
6.33 |
6.66 |
15.33 |
15.00 |
15.66 |
11.66 |
11.66 |
17.67 |
7.66 |
6.66 |
|
12 |
C. jejuni |
6.66 |
6.33 |
12.33 |
12.33 |
12.33 |
11.33 |
0 |
16.00 |
0 |
6.00 |
|
13 |
H. pylori |
6.33 |
0 |
11.33 |
11.66 |
14.66 |
12.00 |
6.00 |
13.33 |
7.00 |
10.33 |
* includes well
size of 5 mm diameter
A1=4-aminobenzonitrile, A 2= 4-aminobenzonitrile, A3=
3-floroaniline, A 4= 4-bromoaniline, A5= 3-cloroaniline, A6= 3-nitroaniline, A7
= 2-nitroaniline, A8 = 2-cloroaniline, A9= 4-nitroaniline, 10 = Aniline
Minimum inhibitory concentration and
Minimum Bactericidal concentration- The minimum inhibitory concentration of synthesized Curcumin
pyrazole triazine conjugate A5 having a best antimicrobial activity was
determined as depicted in Table 2 by resazurin based micro broth dilution
method. The drugs were taken in the concentration ranges as 512, 256, 128, 64
and 32 µg/ml. Among gram-negative bacteria, the best MIC was observed in the
case of P. aeruginosa 32
µg/ml as compared to H.
pylori 64 µg/ml
and E. coli, S. dysentriae with MIC value
128 µg/ml. Among Gram-positive
bacteria, S. aureus was found to have the lowest MIC that is 32 µg/ml than B. subtilis and L. monocytogenes showed MIC at
64 µg/ml and the highest value 128 µg/ml was observed in the case of C. perfringens. Both
gram-negative bacteria P. aeruginosa and
gram-positive bacteria S. aureus were found to have good minimum
bactericidal activity at 128 µg/ml concentration. Except for E. coli with an MBC value of 512 µg/ml all
microbes such as B. subtilis, S. dysentriae, L. monocytogenes and H. pylori reported with
highest MBC value of concentration 256 µg/ml.
Table 2: MIC and MBC of bacterial pathogens
|
S.No. |
Organisms |
MIC (µg/ml) |
MBC (µg/ml) |
|
1 |
E. coli |
128 |
512 |
|
2 |
S. aureus |
32 |
128 |
|
3 |
B. subtilis |
64 |
256 |
|
4 |
S.
dysenteriae |
128 |
256 |
|
5 |
C. perfringens |
128 |
256 |
|
6 |
L.
monocytogenes |
64 |
256 |
|
7 |
H.
pylori |
64 |
256 |
|
8 |
P. aeruginosa |
32 |
128 |
Antifungal activity of synthesized curcumin derivatives- Varied curcumin pyrazole triazine conjugates were tested
for antifungal activity using the agar well diffusion process, shown in Table 3
in accordance with CLSI. All synthesized compounds were found to be
resistant to the pathogenic mold A. fumigatus. Only four drugs
displayed antifungal efficacy against pathogenic yeast Candida albicans with compound A5 having
the most potent zone of inhibition (21 mm) and compound A4 having the least
potent zone of inhibition (14 mm). As a consequence of the findings, chlorine-containing
aniline derivatives were discovered to be the most potent antifungal agent. The
presence of a chlorine atom in meta-position increased its antifungal effect.
In comparison to these halogens, fluorine at ortho-position and bromine at para-position
yielded important effects, but not as much as the chlorine-containing aniline
derivatives of curcumin pyrazole triazine conjugates.
Table 3: Antifungal activity of curcumin
pyrazole triazine conjugates
|
Compounds |
Zone of inhibition (mm diameter) * |
|
|
C. albicans |
A. fumigatus |
|
|
A1 |
0 |
0 |
|
A2 |
0 |
0 |
|
A3 |
15 |
0 |
|
A4 |
14 |
0 |
|
A5 |
21 |
0 |
|
A6 |
0 |
0 |
|
A7 |
0 |
0 |
|
A8 |
17 |
0 |
|
A9 |
0 |
0 |
|
A10 |
0 |
0 |
* includes the
well size of 5 mm diameter
A1=4-aminobenzonitrile, A 2=
4-aminobenzonitrile, A3= 3-floroaniline, A 4 = 4-bromoaniline, A5 =
3-cloroaniline, A6= 3-nitroaniline, A7 = 2-nitroaniline, A8 = 2-cloroaniline,
A9= 4-nitroaniline, 10 = Aniline
DISCUSSION-
Concerned about the ongoing emergence of
multidrug-resistant bacterial and fungal disease, there is an urgent need for
the development of new antimicrobial agents [22,23]. Even newly
developed antibiotics are ineffective against these MDRs [24]. In
this regard, we proposed unique curcumin pyrazole triazine conjugates that have
been found to have antibacterial and antifungal action. Nitrogen-containing
heterocyclic compounds with higher binding affinities to biological receptors
have piqued the interest of many researchers as compounds to use as conjugating
moieties [25]. As a result of its increased importance in biological
studies, particularly for antibacterial [26] and antifungal activity
[19,27]. A nitrogen-containing heterocyclic compound with
good biological activities known as 2, 4, 6-trichloro-1, 3, 5-triazine has been
used as a conjugating agent with curcumin. For verification of synthesized
curcumin pyrazole triazine conjugates, their structures were characterized by
different spectroscopic methods such as FTIR, FT NMR (1H NMR and 13C
NMR) and Mass Spectrometry.
Thereafter synthesized curcumin pyrazole triazine conjugates were evaluated for
the antibacterial and antifungal activity that resulted in significant outcomes
against both fungal and bacterial pathogens.
Against
both types of pathogens compound A5 containing 3-chloroaniline moiety was found
to possess the most potent antimicrobial activity. Among six gram-positive
bacteria only four L. monocytogenes, S. aureus, B. subtilis and C. perfringens were found to be
most susceptible to compound A5 having a zone of inhibition 14, 26, 27, and 25
mm and MIC values of 64, 32, 64 and 128 µg/ml. Among seven gram-negative
bacterial E. coli, S. dysenteriae, H. pylori, P. aeruginosa was reported with the best zone of
inhibition 18, 16, 15, and 28 mm and MIC values with 128, 128, 64 and 32µg/ml.
Regarding antifungal agent similar compound A5 was found to be the most
potent against C. albicans with zone of inhibition of 21 mm. All
synthesized compounds were found to be resistant to the pathogenic A.
fumigatus. According to these findings, curcumin pyrazole triazine
conjugates containing halogen derived anilines such as Cl, Br, and F can bear a
biologically significant hybrid molecule, which could address a lead compound
for various targets and may provide the possibility of reducing multidrug
resistance.
CONCLUSIONS- Novel
curcumin pyrazole triazine conjugates were synthesized bearing covalent linkage
with suitable ligands to enhance its biological activity and characterized by
the FTIR 1HNMR, 13C NMR and Mass spectrometry. The
investigation of antibacterial and antifungal screening data revealed that all
the Curcumin derivatives bearing halogen moiety showed moderate to good
bacterial and fungal inhibition. Significant results were obtained in the case
of both gram-positive bacteria Bacillus subtilis with the zone of inhibition 28 mm and MIC 32 µg/ml and
gram-negative bacteria Pseudomonas aeruginosa with the zone of
inhibition 28 mm and MIC 32 µg/ml. Among pathogenic fungi, Candida albicans
was found to be most susceptible to compound A5. The pharmaceutically important
hybrid molecules can be used in future to cure bacterial and fungal diseases.
ACKNOWLEDGEMENTS- The
authors are thankful to the Department of Pharmaceutical Sciences and the
Department of Industrial Microbiology, Sam Higginbottom University of
Agriculture, Technology and Sciences for providing necessary research facilities to carrying out experiments.
CONTRIBUTION OF AUTHORS
Research concept- Dr. Ebenezer Jeyakumar, Dr.
Rubina Lawrence
Research design- Dr. Ebenezer Jeyakumar, Dr.
Uday Pratap Singh
Supervision- Dr. Ebenezer Jeyakumar
Materials-Anjali
Data collection-Anjali
Data analysis and
Interpretation- Dr. Uday Pratap Singh, Anjali
Literature search- Anjali
Writing article- Anjali
Critical review- Dr. P. Malairajan
Article editing- Anjali
Final
approval- Dr.
Ebenezer Jeyakumar
REFERENCES
1.
Tanwar J, Das S, Fatima Z,
Hameed S. Multidrug Resistance: An
Emerging Crisis. Inter Pers Inf Dis., 2014; pp. 1-7.
2. Roca I, Akova M, Baquero F, Carlet J,
Cavaleri M, et al. The global threat of antimicrobial resistance: science for
intervention. New Microbes New Infect., 2015; 16(6): 22-9.
3. Noval M, Banoub M, Claeys KC, Heil E.
The Battle Is on: New Beta-Lactams for the Treatment of Multidrug-Resistant
Gram-Negative Organisms. Cur Infect Dis Rep., 2020; 22(1): 1.
4. Reddy P, Chadaga S, Noskin GA.
Antibiotic Considerations in the Treatment of Multidrug-Resistant (MDR)
Pathogens: A Case-Based Review. J Hosp
Med., 2009; 4(6):
E8-15.
5.
Nguyen L, Garcia J,
Gruenberg K, Conan, MacDougall. Multidrug-Resistant Pseudomonas Infections:
Hard to Treat, But Hope on the Horizon?. Curr Infect Dis Rep., 2018; 20(8): 23.
6.
Kumar
A, Jaitak V. Natural products as multidrug resistance modulators in cancer. Eur
J Med Chem., 2019; 176: 268-91.
7. Su T, Qiu Y, Hua X, Ye B, Luo H, et al. Novel opportunity to Reverse
antibiotic resistance: To explore traditional chinese medicine with potential
activity against antibiotics-resistance bacteria. Front Microbiol., 2020; 11:
61-70.
8. Chew J, Peh S, Yeang TS. Non-microbial
Natural Products That Inhibit Drug- Resistant Staphylococcus aureus. Intechopen., 2018; pp. 1-31.
9. Corta A, Ozbena T. Natural product
modulators to overcome multidrug resistance in cancer. Nutr Cancer, 2015; pp.
1-13.
10. Khor P Y, Aluwi MFFM, Rullah K, Lam KW.
Insights on the synthesis of asymmetric curcumin derivatives and their
biological activities. Eur J Med Chem., 2019; 1; 183: 111704.
11. Hewlings DJ, Kalman DS. Curcumin: A
Review of Its’ Effects on Human Health. Foods, 2017; 6(10): 92.
12. Lahlou M. The Success of Natural
Products in Drug Discovery. J Pharm Pharmcol., 2013; 4: 17-31.
13.
Aggarwal
BB, Gupta SC, Sung B. Curcumin: an orally bioavailable blocker of TNF and other
pro-inflammatory biomarkers. Br J
Pharmcol., 2013; 169: 1672–92.
14.
Carlos AM, Nano MLT,
Rosatella AA. Synthesis of 2, 4, 6-trisubstituted 1, 3, 5-triazines. Mole., 2006; 81-102.
15. Sarmah KN, Sarmah NK, Kurmi KB, Patel
TV. Synthesis of novel derivatives containing s-triazine moiety as potential
antibacterial agents. Arch Appl Sci Res., 2012; 4: 805-08.
16.
Bhat HR, Pandey Pk, Ghosh
SK, Singh UP. Development of 4-aminoquinolone-1, 3, 5-triazine conjugates as
potent antibacterial agent through facile synthetic route. Med Chem Res., 2013;
28: 8-16.
17.
Al-Khodir FAI, Al-Warti T,
Abumelha HMA, Al-Issa SA. Synthesis, chemical and biological investigations of
new Ru(III) and Se(IV) complexes containing 1,3,5-triazine chelating derivatives.
J Mol Struct., 2018; 795-808.
18.
Solankee A, Kapadia K,
Sokovic AM, Dotchinova I, Geronikaki A. Synthesis of some new s-triazine based
chalcones and their derivatives as potent antimicrobial agents. Eur J Med
chem., 2010; 510-18.
19.
Singh UP, Pathak M, Dubey V,
Bhat HR, Gahtori, et al. Design, synthesis, antibacterial activity and
molecular docking studies of novel hybrids 1, 3-thiazine-1, 3, 5-triazine
derivatives as bacterial translational
inhibitor. Chem Biol Drug Des., 2012; 80: 572–583.
20.
Andrews J. Determination of
minimum inhibitory concentrations. J Antimicrobe Chemotherap., 2001; 48: 5-16.
21. Mariita R, Ogol C, Oguge N, Okeno P. Methanol extract of three
medicinal plants from Samburu in Northern Kenya show significant
antimycobacterial, antibacterial and antifungal properties. J Med Plant Res., 2011; 5(1): 54-64.
22.
Srivastava P, Shukla M, Kaul
G, Chopra S, Patra AK. Rationally designed curcumin based ruthenium (II)
antimicrobials effective against drug-resistant Staphylococcus aureus. Dalton Trans., 2019; 48: 11822–28.
23.
Vivas R, Andrea A, Barbosa
T, Santana S, Dolabela, Jain S.
Multidrug-Resistant Bacteria and Alternative Methods to Control Them: An
Overview. Microbe Drug Resist., 2019; 25(6): 890-908.
24.
Che CT, Zhang H. Plant
Natural Products for Human Health. Int J
Mol Sci., 2019; (4): 830.
25.
Desai NC, Malwana AH, Senta
RD. Synthesis, characterization and antimicrobial activity of some new
4-(40(2-isoniocotinoyl
hydrazinyl)-6-((aryl)amino)-1,3,5-triazin-2-ylamino)-N-(pyrimidin-2-yl)
benzenesulf onamides. Indian J Chem., 2013; 5(1): 21-7.
26.
Patel PK, Patel RV, Mahajan
DH, Parikh PA, Mehta GN, et al. Design, synthesis, characterization and in
vitro antimicrobial activity of novel trisubstituted s-triazines. Med Chem
Res., 2012; 3182-94.
27. Duan Y, Li K, Wang H, Wu T, Zhao Y, et al. Preparation and
evaluation of Curcumin grafted hyaluronic acid modified pollan polymers as a
functional wound dressing material. Carbohydr Polym., 2020; 238: 116195.